
nrStar™ Human functional LncRNA PCR 芯片
产品名称: nrStar™ Human functional LncRNA PCR 芯片
英文名称:
产品编号: Arraystar
产品价格: 0
产品产地: 美国
品牌商标: Arraystar
更新时间: null
使用范围: null
- 联系人 :
- 地址 : 上海市宜山路700号C2栋3楼 (200233)
- 邮编 : 200233
- 所在区域 : 上海
- 电话 : 00 点击查看
- 传真 : 点击查看
- 邮箱 : wangyanling@kangchen.com.cn;market@kangchen.com.cn
nrStar™ Human functional LncRNA PCR 芯片
Arraystar公司最新推出的nrStar™ Human Functional LncRNA PCR Array,可同时检测372个功能已知或疾病相关的金标准lncRNA。这些lncRNA均精心挑选于科学文献和最新数据库。与大部分功能未知、序列残缺的lncRNA相比,产品中的lncRNA特征明确、信息完善。Arraystar为这些lncRNA提供了详细的注释信息,包括生物学功能(干细胞分化、胚胎发育、心血管发育、造血系统与免疫、内分泌系统等)和作用机制[1](基因表达调控、表观遗传、基因组印记、染色体修饰等)等(图1)。lncRNA表达失调还与和神经退行性疾病、心血管疾病[2]、糖尿病和癌症[3]等病理特征紧密相关。另外,LncRNA在表达分布和组织/疾病特异性上高于mRNA,已有不少已知或未知功能的lncRNA被认为具有分子标志物潜能(图2)。比如,PCA3已通过FDA认证,应用于前列腺癌的临床诊断。
Arraystar功能性lncRNA PCR芯片不仅含有最佳的内容,而且具有最优的技术表现。通过靶向特定的外显子或可变剪接位点,每对lncRNA引物可以检测到特定的转录本,从而区分同一lncRNA基因的不同转录本亚型。“机智”的cDNA合成策略保证了从完整或降解的RNA样本检测多聚腺苷酸尾和非多聚腺苷酸尾lncRNA的有效性。严格的外参、内参设置以及可信度高的均一化策略,保证了高质量的qPCR结果及其精确定量,可充分满足临床诊断应用或分子标志物鉴定所需。
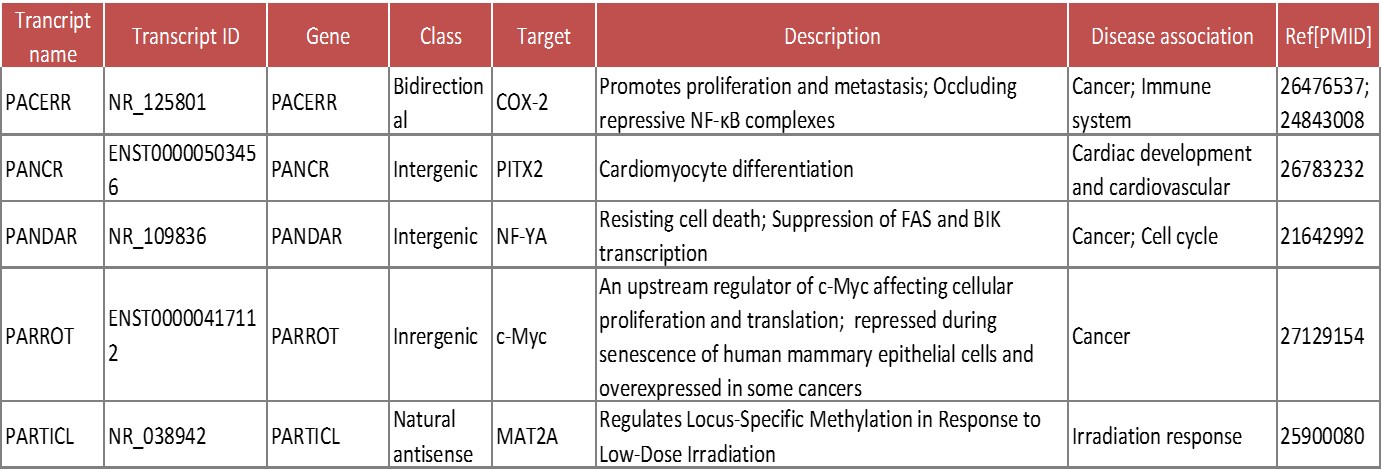
表一:金标准lncRNA注释信息部分截图
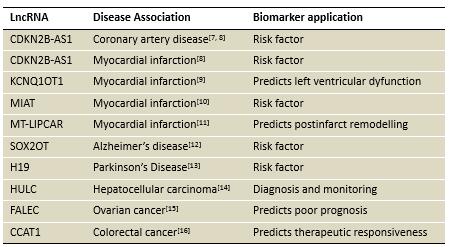
表二:部分已报道的具有分子标志物潜能的lncRNA列表
产品列表
|
芯片名称 |
规格 |
描述 |
|
nrStar™ Human functional LncRNA PCR Array |
1 plate |
包含372个与生物学功能和疾病紧密关联的已知功能lncRNA转录本 |
芯片特点
● 覆盖最新最全的已知功能lncRNA:从最新数据库和科研文献中收集了372个与生物学过程和疾病明确相关的LncRNA转录本。PCR芯片包含目前市场上最新最全的已知功能LncRNA。
● 强效检测:无论样本是否降解,lncRNA是否有Poly(A)尾,都可被Arraystar“机智”的流程所检测,得到高质量的数据结果。
● 最可靠和最精准的lncRNA定量检测:通过使用一系列严格的外参和内参,芯片中的每个LncRNA转录本都能被准确和可靠的定量检测。与常规使用一个内参基因不同,Arraystar公司产品使用生物信息学方法从多个候选内参基因中挑选出最适的内参基因进行数据均一化。
● 转录本特异性检测:通过靶向特异性的外显子或可变剪接点,芯片中的PCR引物准确有效的检测每个LncRNA转录本。
● 稳定有效的PCR引物:所有LncRNA引物均经过精心优化,并在多种细胞和组织中通过验证。
● 简单便捷:384孔板,即拆即用。只需将反转好的cDNA与qPCR Master Mix混合,加入到384孔板,即可进行qPCR反应。所有流程可在4小时内完成。
工作流程
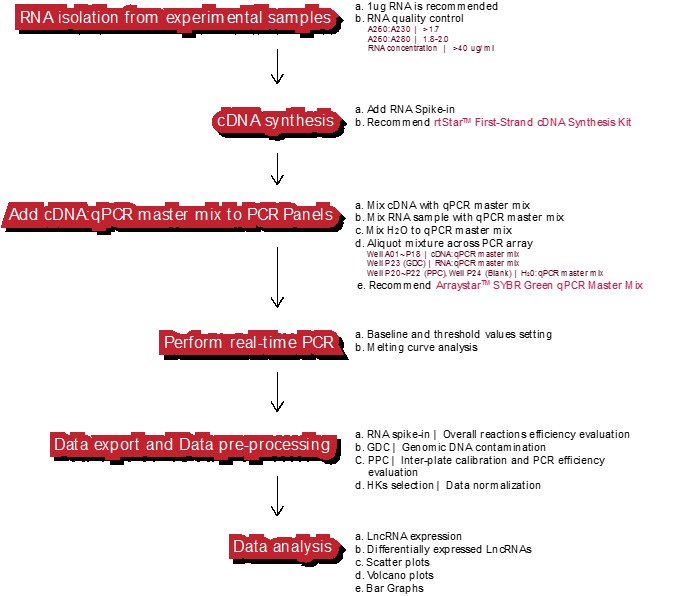
适用实时定量荧光PCR仪:
ABI ViiA™ 7,ABI 7500 & ABI 7500 FAST,ABI 7900HT,ABI QuantStudio™ 6 Flex Real-Time PCR system,ABI QuantStudio™ 7 Flex Real-Time PCR system,ABI QuantStudio™ 12K Flex Real-Time PCR System,Bio-Rad CFX384,Bio-Rad iCycler & iQ Real-Time PCR Systems,Eppendorf Realplex,QIAGEN Rotor Gene Q 100,Roche LightCycler 480,Stratagene Mx3000,Roche LightCycler 480
数据库
|
Cancer [222]: |
|
Neurodegenerative diseases [42]: |
|
Cardiovascular diseases [44]: |
|
Kidney diseases [17]: |
|
Diabetes [10]: |
|
Immune system [15]: |
|
Cell Cycle [11]: |
|
Lipid metabolism and adipogenesis [8]: |
注:后缀(-1, -2, -3 …) 代表lncRNA基因的不同转录本。
参考文献
[1] Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nature reviews Genetics 2014;15:7-21.
[2] Devaux Y, Zangrando J, Schroen B, Creemers EE, Pedrazzini T, Chang CP, et al. Long noncoding RNAs in cardiac development and ageing. Nature reviews Cardiology 2015;12:415-25.
[3] Satpathy AT, Chang HY. Long noncoding RNA in hematopoiesis and immunity. Immunity 2015;42:792-804.
[4] Knoll M, Lodish HF, Sun L. Long non-coding RNAs as regulators of the endocrine system. Nature reviews Endocrinology 2015;11:151-60.
[5] Lorenzen JM, Thum T. Long noncoding RNAs in kidney and cardiovascular diseases. Nature reviews Nephrology 2016;12:360-73.
[6] Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer cell 2016;29:452-63.
[7] Consortium CAD, Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nature genetics 2013;45:25-33.
[8] Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, et al. Genomewide association analysis of coronary artery disease. The New England journal of medicine 2007;357:443-53.
[9] Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circulation research 2014;115:668-77.
[10] Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. Journal of human genetics 2006;51:1087-99.
[11] Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circulation research 2014;114:1569-75.
[12] Arisi I, D'Onofrio M, Brandi R, Felsani A, Capsoni S, Drovandi G, et al. Gene expression biomarkers in the brain of a mouse model for Alzheimer's disease: mining of microarray data by logic classification and feature selection. Journal of Alzheimer's disease : JAD 2011;24:721-38.
[13] Kraus TF, Haider M, Spanner J, Steinmaurer M, Dietinger V, Kretzschmar HA. Altered Long Noncoding RNA Expression Precedes the Course of Parkinson's Disease-a Preliminary Report. Molecular neurobiology 2016.
[14] Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. BioMed research international 2013;2013:136106.
[15] Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer cell 2014;26:344-57.
[16] McCleland ML, Mesh K, Lorenzana E, Chopra VS, Segal E, Watanabe C, et al. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. The Journal of clinical investigation 2016;126:639-52.
上海康成生物工程有限公司
地址:上海市虹漕路421号63号楼2楼
全国免费热线:400-886-5058;800-820-5058
电话:021-64451989
传真:021-64452021
网址:www.kangchen.com.cn
